Tutorial 3.1 Gene Ontology enrichment analysis¶
Analyse the conservation patterns of genes involved in different biological processes¶
We will use a data set containing the Conservation Score for all human genes to their closest ortholog in 15 other organisms and we will reproduce the results described in Lopez-Bigas et al 2008 .
Files needed¶
hsapiens_cs_orthologs_EnsV42.cdm.gz which contains the conservation score of all human genes to their closest ortholog in 15 other organisms.
EnsGenesV42_GOprocess.tcm.gz which is the module file mapping Ensembl genes (V42) to Gene Ontology Biological Process terms.
Perform an enrichment analysis with Gitools¶
See this chapter for details on how to perform enrichment analysis
Select hsapiens_cs_orthologs_EnsV42.cdm.gz as data file
Select the option “Filter out rows for which no information appears in the modules”
Select the GO annotations file as module file: file:EnsGenesV42_GOprocess.tcm.gz .
Select zscore statistical test. Write 100 in sampling size for a quick test of the analysis. To get a definitive result run the analysis with 10000, however take into account that in this case the anlysis will take long time to finish. Leave estimator and multiple test correction as default.
Give a name to the analysis. Select a directory where to save it and click Finish.
If you have a memory problem, see memory configuration in ( Installation ) to increase the memory allocated to run Gitools.
Filter the rows of the matrix with this list of GO terms ( GOprocess_shortlist.txt ). Go to Data>Filter>Filter by label.
Explore the results¶
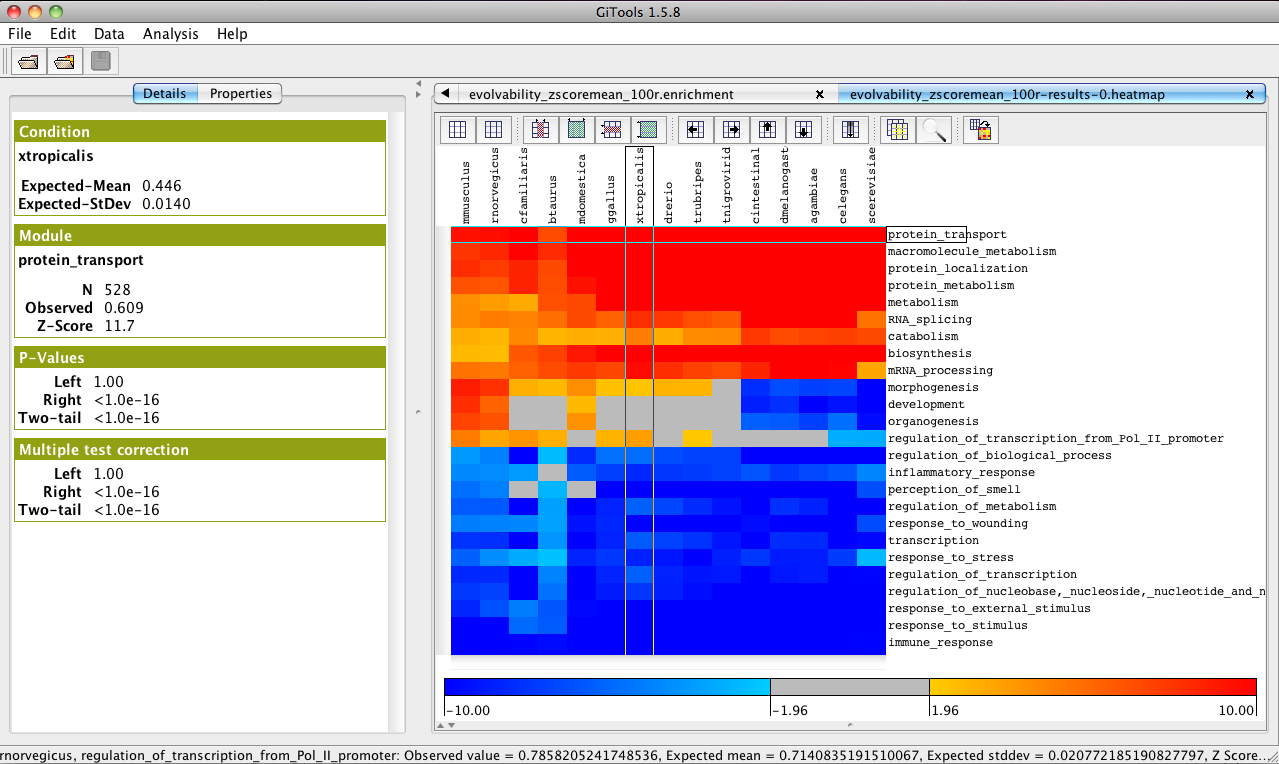
In the analysis details tab, click on “heatmap” under “Results” to view the heatmap of the results.
Change the colour scale to z-score scale in the Settings tab under “scale”.
Filter significance by Corrected two-tail p-value by checking the box below.